Another explanation could be that Chloroquine Phosphate TcSub2 is related to transcription of specific mRNAs as consequence of its role in regulation of gene expression. To further investigate the function of TcSub2, we attempted to knockout the TcSub2 gene to obtain phenotypic Chlorhexidine hydrochloride alterations related to mRNA export. However, the null knockout lineages of T. cruzi were not viable. Studies show that it is an essential protein in other eukaryotes because the knockdown of Sub2 homologs inhibits cell growth. Unfortunately, knockout of a single TcSub2 allele was not enough to affect morphology and cell growth. Although the single-allele TcSub2 knockouts had around 50% less TcSub2 protein, this did not alter mRNA export. It is therefore necessary to apply a regulated system to completely silence TcSub2 expression to get more information regarding to its function in T. cruzi. The absence of approaches of inducible silencing, such as RNA interference, in T. cruzi preclude suitable studies for this. On the other hand, T. bruceii is a suitable model to study essential genes using inducible system based on RNAi in trypanosomes. Taking in account that Sub2 is highly conserved in trypanosomes, we decided to use T. bruceii to study the function of this protein. The data obtained demonstrated that it is really an essential protein since its knockdown inhibits the cellular growth. Similar to other eukaryotes, the silencing of TbSub2 expression caused 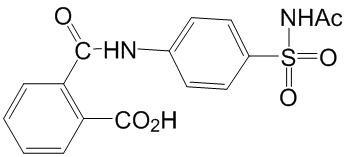 translation inhibition and robust accumulation of bulk polyadenylated mRNAs in the nucleoplasm. These phenotypes are also observed when the expression of homologs is silenced in metazoan.Interestingly, we also observed an arrest of cell cycle in G2 phase, mainly after 72 hours of RNAi induction when the level of the proteins was reduced by 70%. In the same way, knockdown of UAP56, the homolog in humans, resulted in mitotic defects. Taking together, the trypanosome protein has similar characteristics to its orthologs in higher eukaryotes, possibly reflecting a similar function in these parasites. In the context of nuclear mRNA metabolism in trypanosomatids, nucleocytoplasmic transport is not well understood and data in this area will lend further insights into regulation of gene expression in these parasites. Herein, we identified a novel trypanosome protein that it is related to nuclear mRNA metabolism and proposed that the Tryp-Sub2 is a component of mRNA transcription/export pathway in these parasites. Whereas approximately half of CLL patients have an indolent clinical course that does not require treatment for many years, a more aggressive clinical course that necessitates treatment within a few years is diagnosed for the other half. These differences in clinical course correlate with molecular markers, including the mutational status of the surface immunoglobulins and the expression of intracellular tyrosine kinase ZAP-70. Despite a typically lower expression of CD20 compared to normal B cells, the combination of fludarabine and cyclophosphamide with the chimeric mouse/human anti-CD20 IgG1 monoclonal antibody rituximab has become standard first-line treatment that was approved by the Food and Drug Administration in 2010. In addition, alemtuzumab, a humanized anti-CD52 IgG1 mAb, was FDA-approved in 2001 as single agent for CLL therapy. Alemtuzumab is frequently used for second-line treatment, but is ineffective in CLL patients with bulky lymphadenopathy. In 2009, fully human IgG1 mAb ofatumumab, which binds to a CD20 epitope different from rituximab, was FDA-approved for treating CLL patients refractory to fludarabine and alemtuzumab.
translation inhibition and robust accumulation of bulk polyadenylated mRNAs in the nucleoplasm. These phenotypes are also observed when the expression of homologs is silenced in metazoan.Interestingly, we also observed an arrest of cell cycle in G2 phase, mainly after 72 hours of RNAi induction when the level of the proteins was reduced by 70%. In the same way, knockdown of UAP56, the homolog in humans, resulted in mitotic defects. Taking together, the trypanosome protein has similar characteristics to its orthologs in higher eukaryotes, possibly reflecting a similar function in these parasites. In the context of nuclear mRNA metabolism in trypanosomatids, nucleocytoplasmic transport is not well understood and data in this area will lend further insights into regulation of gene expression in these parasites. Herein, we identified a novel trypanosome protein that it is related to nuclear mRNA metabolism and proposed that the Tryp-Sub2 is a component of mRNA transcription/export pathway in these parasites. Whereas approximately half of CLL patients have an indolent clinical course that does not require treatment for many years, a more aggressive clinical course that necessitates treatment within a few years is diagnosed for the other half. These differences in clinical course correlate with molecular markers, including the mutational status of the surface immunoglobulins and the expression of intracellular tyrosine kinase ZAP-70. Despite a typically lower expression of CD20 compared to normal B cells, the combination of fludarabine and cyclophosphamide with the chimeric mouse/human anti-CD20 IgG1 monoclonal antibody rituximab has become standard first-line treatment that was approved by the Food and Drug Administration in 2010. In addition, alemtuzumab, a humanized anti-CD52 IgG1 mAb, was FDA-approved in 2001 as single agent for CLL therapy. Alemtuzumab is frequently used for second-line treatment, but is ineffective in CLL patients with bulky lymphadenopathy. In 2009, fully human IgG1 mAb ofatumumab, which binds to a CD20 epitope different from rituximab, was FDA-approved for treating CLL patients refractory to fludarabine and alemtuzumab.
Identification of highly variable and approach to predict drugtarget interactions
Which is a new contribution in the field of drug discovery and development. As well known, RF introduces two sources of randomness into the trees: random training set and random input vectors. Each tree is grown using a bootstrap sample of training data at each node, with best split chosen from random sample of mtry variables instead of all variables. The Bagging method causes the low bias and high variance of the unpruned trees, but the variance is reduced by averaging the bootstrapped trees. Because each node is split using the best among a subset of predictors Albaspidin-AA randomly chosen at that node, this strategy turns out to perform very well compared with many other classifiers such as ANN and SVM, and is more robustness against overfitting. However, it should be also pointed out that SVM allows us to use a tensor product space, with no extra calculation time with respect to the joint space, and versatile choice of similarity measures for proteins and ligands, which is a clear advantage of SVM with respect to this proposed RF approach. As shown in the Results section, the proposed method enables to identify the unrelated proteins that may share structurally similar pockets in the 3D space, the advantage effect of which is derived from the learning ligand similarities. If the unrelated receptors do not possess similar ligands in the learning dataset, then the protein encoding plays a key role for the unrelated targets identification, which have been well applied for the prediction of protein structural and functional classes, protein-protein interactions and subcellular locations. Particularly, most descriptors of this encoding such as Composition of Solvent Accessibility and Distribution of Charge are highly useful for representing and distinguishing interaction profiles, which is essential for the successful application of statistical learning methods in predicting the interaction profiles between drugs and targets. Although this encoding can effectively describe the interaction profiles of the whole protein, it is limited by the description and extraction of the 3D structure features of the ligand-binding domains. Therefore, obtaining optimal protein encoding method will require further research. The main advantages of our proposed approach are summarized as follows: 1) In building models, no explicit procedure is needed to select the information shared among all drug-target recognitions. 2) Our system is suitable for simultaneously screening huge numbers of drug candidates and candidate targets from a systematic level; 3) Compared with the structure-based simulation methods, this approach is not limited by the 3D structure data of targets; 4) The structure similarity of 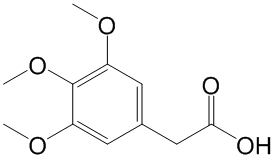 a chemical with ligands that bind to proteins which this compound is not known to bind can be used by our method to discover unknown Catharanthine sulfate activities. 5) The system is able to identify those ligands/receptors that exhibit scaffold hopping/low sequence similarity; and 6) this approach can aid in discovery of multi-target drugs by recognizing the group of proteins targeted by a particular ligand. Based on all these, we conclude that the present systematic biology-based “integrationist mindset” is appropriate for modeling and understanding complex drug-target interaction networks. This perspective could devote to the recapturing of known small molecules and the explication of mechanisms of drug side effects, and finally is anticipated to help fill in the existing gap between chemical genomics and network pharmacology. Massive UDPS analysis enabled sensitive baseline detection of minor viral populations associated with NA resistance.
a chemical with ligands that bind to proteins which this compound is not known to bind can be used by our method to discover unknown Catharanthine sulfate activities. 5) The system is able to identify those ligands/receptors that exhibit scaffold hopping/low sequence similarity; and 6) this approach can aid in discovery of multi-target drugs by recognizing the group of proteins targeted by a particular ligand. Based on all these, we conclude that the present systematic biology-based “integrationist mindset” is appropriate for modeling and understanding complex drug-target interaction networks. This perspective could devote to the recapturing of known small molecules and the explication of mechanisms of drug side effects, and finally is anticipated to help fill in the existing gap between chemical genomics and network pharmacology. Massive UDPS analysis enabled sensitive baseline detection of minor viral populations associated with NA resistance.
Selective inability of hNGF R100 protein to activate nociception while retaining the neurotrophic properties
Indeed, the binding affinity of hNGFR100 mutants to p75NTR is strongly reduced, while that for TrkA remains unchanged. Despite the identical TrkA binding affinity, hNGFR100 mutants differ from hNGF in their ability to activate downstream TrkAdependent signalling pathways, with a notable selective reduction in the ability to activate PLC�C1c and Erks. Mechlorethamine hydrochloride Notably, hNGFR100 mutants do not sensitize nociceptors and fail to induce pain in vivo. Thus, hNGFR100 mutants may share some properties of other NGF mutants described to be selective TrkA agonists with respect to p75NTR, but have the additional property of selectively modulating a subset of the signaling pathways downstream of TrkA, and to be proven as less potent nociception activators. On the basis of these properties, the hNGFP61S/R100E therapeutic candidate was designed, aimed at optimizing the therapeutic window for NGF. The characterization of the hNGFP61S/R100E double mutant is provided in this paper. Moreover, until recently no study addressed the issue whether administration of NGF, or of NGF-related molecules, might affect the neurodegeneration in 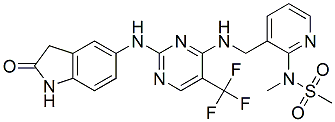 FAD related APP mouse models. We show that hNGFP61S/R100E is characterized by a reduced ability to activate PLC�C1c, Erks and c-jun, providing a molecular explanation for the reduced pro-nociceptive activity, observed in a mechanical allodynia pain model. hNGFP61S/R100E showed no difference with respect the hNGF counterparts in a number of different survival, differentiation and proliferation cellular assays. Thus, hNGFR100P61S/R100E maintains the neurotrophic signaling stream unchanged, while showing a reduced signaling involved in nociceptor sensitization and pain perception. Notably, hNGFP61S/R100E is very effective to rescue or prevent neurodegeneration and learning and memory deficits in mouse models, including an APP-based model, in which the neurodegeneration is not directly caused by NGF inhibition. We observed a higher effectiveness of hNGFP61S/R100E, with respect to hNGFR100E, in rescuing the neurodegeneration in the AD11 model, as well as a slight difference between the R100E and the P61S/R100E NGF proteins in nociceptive sensitization assays, most likely involving a structural cross-talk between S61 and R100 residues. Thus, the P61S substitution, which per se does not determine any measurable influence, Cinoxacin compared to wild type NGF,, could exert a long distance influence on the residue at position 100. In all the available 3D structures of hNGF complexes loop 58�C68 is only partially resolved. Indeed the region 61�C66, including the residue P61, is structurally not defined. On the contrary, in the crystal structures of mouse NGF the homologous loop, including S61, is structurally well defined. Thus, the 58�C68 loop in hNGF has likely a higher intrinsic conformational flexibility compared to mNGF. This loop is structurally positioned over the C58�CC108 and C68�CC110 cysteine knot, and it may pose structural constraints on the possible conformations taken by the disulphide bridges of the cysteine knot. The C58�CC108 and C68�CC110 disulphide bridges pair b-strand-2 to b-strand-4. Residue R100 is located at the N-term of b-strand-4. The P61S substitution in hNGF, by rendering the 58�C68 loop structurally similar to that observed in mNGF, might cause a local conformational change in the proximity of the interacting cysteine knot, thereby exerting a long-distance effect on R100, which could explain the observed subtle modifi
FAD related APP mouse models. We show that hNGFP61S/R100E is characterized by a reduced ability to activate PLC�C1c, Erks and c-jun, providing a molecular explanation for the reduced pro-nociceptive activity, observed in a mechanical allodynia pain model. hNGFP61S/R100E showed no difference with respect the hNGF counterparts in a number of different survival, differentiation and proliferation cellular assays. Thus, hNGFR100P61S/R100E maintains the neurotrophic signaling stream unchanged, while showing a reduced signaling involved in nociceptor sensitization and pain perception. Notably, hNGFP61S/R100E is very effective to rescue or prevent neurodegeneration and learning and memory deficits in mouse models, including an APP-based model, in which the neurodegeneration is not directly caused by NGF inhibition. We observed a higher effectiveness of hNGFP61S/R100E, with respect to hNGFR100E, in rescuing the neurodegeneration in the AD11 model, as well as a slight difference between the R100E and the P61S/R100E NGF proteins in nociceptive sensitization assays, most likely involving a structural cross-talk between S61 and R100 residues. Thus, the P61S substitution, which per se does not determine any measurable influence, Cinoxacin compared to wild type NGF,, could exert a long distance influence on the residue at position 100. In all the available 3D structures of hNGF complexes loop 58�C68 is only partially resolved. Indeed the region 61�C66, including the residue P61, is structurally not defined. On the contrary, in the crystal structures of mouse NGF the homologous loop, including S61, is structurally well defined. Thus, the 58�C68 loop in hNGF has likely a higher intrinsic conformational flexibility compared to mNGF. This loop is structurally positioned over the C58�CC108 and C68�CC110 cysteine knot, and it may pose structural constraints on the possible conformations taken by the disulphide bridges of the cysteine knot. The C58�CC108 and C68�CC110 disulphide bridges pair b-strand-2 to b-strand-4. Residue R100 is located at the N-term of b-strand-4. The P61S substitution in hNGF, by rendering the 58�C68 loop structurally similar to that observed in mNGF, might cause a local conformational change in the proximity of the interacting cysteine knot, thereby exerting a long-distance effect on R100, which could explain the observed subtle modifi
Residues involved in bond interactions were investigated as they seemed more effects on enzyme activity
This panel shows an array of bi-variate histograms for each possible pair of the top clique residues, even those that are not neighbors. Residue couplings are not explicit on this figure, which instead shows the initial data on joint Ginsenoside-Ro sequence identity used in the MI calculations. However, by selecting neighboring positions the bivariate histograms can be used to find which residue types are found at each position, and their frequencies. Also shown in Fig. 2B is a contact map for the top clique residues, which highlights the physical proximity of many of top clique residues. Some general observations can be made from Fig. 2B. For example, the top clique residues involve covariation between two or three different residue types. In some cases, the coevolving residue types that occur at a given Chloroquine Phosphate position are quite similar. Others coevolve as very different residue types, such as positions 374 and 430. One trend that might be expected from this data is that the physicochemical characteristics of the residue pair would be conserved at co-varying 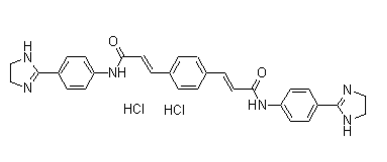 positions, despite changes in residue identity. However, such patterns are not readily apparent for these residues. This may be due to the nature of the structural interactions made by the top clique residues, which are predominantly between atoms in the side chain of one residue and the backbone of the other. The top clique residues fall on both sides of this interface, including residues from domains 3 and 4 of the protein. Indeed, these eight residues form an essentially contiguous residue patch, which spans the width of the domain interface. The remaining residues are found in domains 1, 2, and elsewhere in domain 3, generally near the center of the molecule, but not within the active site cleft. With the exception of K285, which makes ligand contacts in certain enzyme-substrate complexes, none of the top clique residues were of previously implicated functional significance in PMM/PGM. However, the domain 4 interface is a known site of conformational change of the protein, as first observed in crystallographic studies of enzymesubstrate complexes. For comparison with the results of the coevolutionary analysis, Fig. 3C shows the structure of P. aeruginosa PMM/PGM colored according to sequence conservation in the MSA. On this figure, it is clear that the most highly conserved residues cluster in the center of the molecule, in or near the active site. In particular, clusters of conserved residues are found in active site loops in domains 1 and 2 of the protein. These loops include residues involved in the phosphoryl transfer reaction and coordination of the Mg2+ required for enzyme activity. Only a few highly conserved residues are found near the domain 4 interface. Hence, the structural location of highly conserved residues and coevolving residues in PMM/PGM are distinct, although if considered as a group, they would tend to form a contiguous patch in/near the active site, and extending along the domain 4 interface. To investigate the functional role of the coevolving network, a number of the top clique residues were selected for site-directed mutagenesis to alanine. Proline and glycine residues were excluded from consideration, due to possible negative effects on protein folding. Of those remaining, residues were selected based on: i) their location at the interface between domain 4 and the rest of the protein; and, ii) a side chain involved in a hydrogen bond or ion pair interaction with another residue across the interface.
positions, despite changes in residue identity. However, such patterns are not readily apparent for these residues. This may be due to the nature of the structural interactions made by the top clique residues, which are predominantly between atoms in the side chain of one residue and the backbone of the other. The top clique residues fall on both sides of this interface, including residues from domains 3 and 4 of the protein. Indeed, these eight residues form an essentially contiguous residue patch, which spans the width of the domain interface. The remaining residues are found in domains 1, 2, and elsewhere in domain 3, generally near the center of the molecule, but not within the active site cleft. With the exception of K285, which makes ligand contacts in certain enzyme-substrate complexes, none of the top clique residues were of previously implicated functional significance in PMM/PGM. However, the domain 4 interface is a known site of conformational change of the protein, as first observed in crystallographic studies of enzymesubstrate complexes. For comparison with the results of the coevolutionary analysis, Fig. 3C shows the structure of P. aeruginosa PMM/PGM colored according to sequence conservation in the MSA. On this figure, it is clear that the most highly conserved residues cluster in the center of the molecule, in or near the active site. In particular, clusters of conserved residues are found in active site loops in domains 1 and 2 of the protein. These loops include residues involved in the phosphoryl transfer reaction and coordination of the Mg2+ required for enzyme activity. Only a few highly conserved residues are found near the domain 4 interface. Hence, the structural location of highly conserved residues and coevolving residues in PMM/PGM are distinct, although if considered as a group, they would tend to form a contiguous patch in/near the active site, and extending along the domain 4 interface. To investigate the functional role of the coevolving network, a number of the top clique residues were selected for site-directed mutagenesis to alanine. Proline and glycine residues were excluded from consideration, due to possible negative effects on protein folding. Of those remaining, residues were selected based on: i) their location at the interface between domain 4 and the rest of the protein; and, ii) a side chain involved in a hydrogen bond or ion pair interaction with another residue across the interface.
When considered with a necessary accompanying conformational change of the enzyme
The factors governing the conformational flexibility of the protein remains a key area of interest with regard to the function of this enzyme, and others in the superfamily. In the present study, a coevolution analysis was used to examine sequence relationships of enzymes in the PMM/PGM family. Mutual information analyses were used to identify coevolving residue pairs, and “cliques” were calculated using graph theory to find  networks of coevolving residues. We identify a tightly linked network of coevolving residues, most of which localize to the interface between domain 4 and the rest of the protein, which is a wellcharacterized site of conformational variability in the enzyme. This result is in distinct contrast to the highly conserved residues in the protein family that cluster in the active site and tend to be directly involved in catalysis/ligand binding. Furthermore, we report the steady-state kinetic characterization of mutants of residues in the coevolving network, and find a reduction in enzyme specificity relative to Atropine sulfate wild-type, ranging from 45 to less than 10%. Mutation of an interface residue that is not part of the coevolving cluster results in no change in specificity, despite making direct structural contacts with other residues in the network selected for mutation. Double mutants of several key residues in the network show additivity in their effects, suggesting that these residues are not significantly coupled energetically. This study sheds new light on the roles of coevolutionary networks in proteins and has implications for the definition of catalytic residues in enzymes, which, as shown here, can be distant from the active site. The ZNMI approach was chosen as it gave results for our MSA that were better when considering both accuracy and reproducibility relative to the other MI methods, using the definitions from. Accuracy of the various methods was assessed by determining if residue pairs with high scores are, on average, close in tertiary structure, the most commonly used metric for evaluating coevolution analysis. ZNMI was calculated between all ungapped positions in the PMM/PGM alignment as described in Methods. The residue couplings from ZNMI were then subjected to a resampling procedure to eliminate errors due to perturbations of the sequence alignment. For this purpose, ensembles of sub-alignments were utilized in a cross validation approach, as described in. For the present study, only residues with 100% reproducibility as determined by the data resampling were considered for further analysis. This stringent cutoff greatly reduces the possibility of false positives from the MI analysis, and, given the large size of the protein, helps reduce the number of coevolving positions to a LOUREIRIN-B manageable number for analysis. It does, however, have the potential disadvantage of missing significant couplings, a limitation that must be considered in interpretation of results. A matrix of the resampled ZNMI scores for all possible residue position pairs in the PMM/PGM MSA is given in Fig. 1A. Also, for comparison, Fig.1B shows per residue plots of the degree, sequence entropy, and gaps for the MSA. The matrix in Fig. 1A is notably sparse due to application of the 100% reproducibility criterion. Despite this, the number of high-scoring residue pairs from the rZNMI analysis is still quite large, and difficult to assess on an individual basis. In general, however, it can be seen that high-scoring couplings are scattered throughout the sequence, although with higher density in the C-terminal half of the protein.
networks of coevolving residues. We identify a tightly linked network of coevolving residues, most of which localize to the interface between domain 4 and the rest of the protein, which is a wellcharacterized site of conformational variability in the enzyme. This result is in distinct contrast to the highly conserved residues in the protein family that cluster in the active site and tend to be directly involved in catalysis/ligand binding. Furthermore, we report the steady-state kinetic characterization of mutants of residues in the coevolving network, and find a reduction in enzyme specificity relative to Atropine sulfate wild-type, ranging from 45 to less than 10%. Mutation of an interface residue that is not part of the coevolving cluster results in no change in specificity, despite making direct structural contacts with other residues in the network selected for mutation. Double mutants of several key residues in the network show additivity in their effects, suggesting that these residues are not significantly coupled energetically. This study sheds new light on the roles of coevolutionary networks in proteins and has implications for the definition of catalytic residues in enzymes, which, as shown here, can be distant from the active site. The ZNMI approach was chosen as it gave results for our MSA that were better when considering both accuracy and reproducibility relative to the other MI methods, using the definitions from. Accuracy of the various methods was assessed by determining if residue pairs with high scores are, on average, close in tertiary structure, the most commonly used metric for evaluating coevolution analysis. ZNMI was calculated between all ungapped positions in the PMM/PGM alignment as described in Methods. The residue couplings from ZNMI were then subjected to a resampling procedure to eliminate errors due to perturbations of the sequence alignment. For this purpose, ensembles of sub-alignments were utilized in a cross validation approach, as described in. For the present study, only residues with 100% reproducibility as determined by the data resampling were considered for further analysis. This stringent cutoff greatly reduces the possibility of false positives from the MI analysis, and, given the large size of the protein, helps reduce the number of coevolving positions to a LOUREIRIN-B manageable number for analysis. It does, however, have the potential disadvantage of missing significant couplings, a limitation that must be considered in interpretation of results. A matrix of the resampled ZNMI scores for all possible residue position pairs in the PMM/PGM MSA is given in Fig. 1A. Also, for comparison, Fig.1B shows per residue plots of the degree, sequence entropy, and gaps for the MSA. The matrix in Fig. 1A is notably sparse due to application of the 100% reproducibility criterion. Despite this, the number of high-scoring residue pairs from the rZNMI analysis is still quite large, and difficult to assess on an individual basis. In general, however, it can be seen that high-scoring couplings are scattered throughout the sequence, although with higher density in the C-terminal half of the protein.